
In October 2021, while Delta was continuing to exhibit high levels of transmission in the Northern Hemisphere, a large Delta wave was subsiding in southern Africa. During its spread, the Delta variant evolved into multiple sublineages 7, some of which demonstrated signs of having a growth advantage in certain locations 8, prompting speculation that the next VOC to drive a resurgence of infections would probably be derived from Delta. Although the Alpha (B.1.1.7), Beta and Gamma VOCs 2, 5 that emerged during this time disseminated globally and drove epidemic resurgences in many different countries, it was the highly transmissible Delta variant that subsequently displaced all of the other VOCs in most regions of the world 6. These variants exhibited increased transmissibility and/or immune evasion properties that threatened global efforts to control the pandemic. Between October and December 2020, the world witnessed the emergence of the first variants of concern (VOCs). Some variants have spread worldwide and made major contributions to the cyclical infection waves that occur asynchronously in different regions. Since the onset of the COVID-19 pandemic in December 2019, variants of SARS-CoV-2 have emerged repeatedly. Here we describe the genomic profile and early transmission dynamics of Omicron, highlighting the rapid spread in regions with high levels of population immunity.

The Omicron variant is exceptional for carrying over 30 mutations in the spike glycoprotein, which are predicted to influence antibody neutralization and spike function 4. Within three days of the first genome being uploaded, it was designated a variant of concern (Omicron, B.1.1.529) by the World Health Organization and, within three weeks, had been identified in 87 countries. In November 2021, genomic surveillance teams in South Africa and Botswana detected a new SARS-CoV-2 variant associated with a rapid resurgence of infections in Gauteng province, South Africa. The first was associated with a mix of SARS-CoV-2 lineages, while the second and third waves were driven by the Beta (B.1.351) and Delta (B.1.617.2) variants, respectively 1, 2, 3. The SARS-CoV-2 epidemic in southern Africa has been characterized by three distinct waves. Nature volume 603, pages 679–686 ( 2022) Cite this article
#OXFORD NANOPORE MIDNIGHT PROTOCOL WINDOWS#
Installation instructions are available for Linux, Windows and macOS.Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa These tutorials are provided through a Jupyter Labs server running in a docker container.

Analysis of SARS-CoV-2 genome sequence data prepared using ARTIC-based protocols.An introduction to genome assembly and consensus sequence polishing.Review of sequence collections to collate and present summary statistics for QC purposes.File formats that will be encountered during Nanopore sequence analysis (e.g.These tutorials include step-by-step instructions and are packaged with example datasets. A collection of tutorials to introduce best practise solutions to research questions is available through EPI2ME Labs.
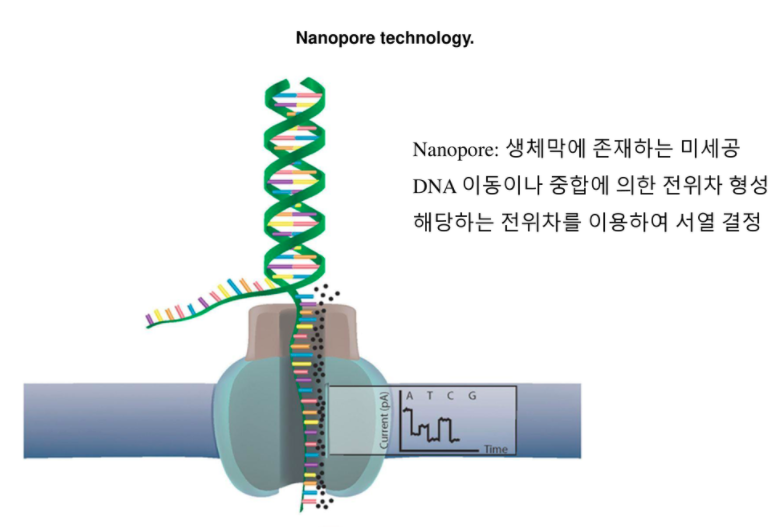
Our research tools are available through our GitHub and are suited for users with bioinformatics experience. Our bioinformatics offerings include research tools, our EPI2ME Labs and EPI2ME products.


 0 kommentar(er)
0 kommentar(er)
